What Name Is Given to the Collection of Traits Exhibited by an Organism?
| | Await upwards phenotype in Wiktionary, the gratuitous lexicon. |

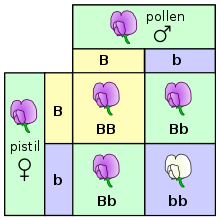
Here the relation betwixt genotype and phenotype is illustrated, using a Punnett square, for the grapheme of petal colour in pea plants. The letters B and b correspond genes for color, and the pictures show the resultant phenotypes. This shows how multiple genotypes (BB and Bb) may yield the same phenotype (majestic petals).
In genetics, the phenotype (from Ancient Greek φαίνω ( phaínō ) 'to appear, show, shine', and τύπος ( túpos ) 'marking, type') is the set of observable characteristics or traits of an organism.[i] [ii] The term covers the organism'due south morphology or physical class and construction, its developmental processes, its biochemical and physiological properties, its beliefs, and the products of behavior. An organism'due south phenotype results from two basic factors: the expression of an organism'south genetic code, or its genotype, and the influence of environmental factors. Both factors may interact, farther affecting phenotype. When two or more conspicuously different phenotypes exist in the same population of a species, the species is called polymorphic. A well-documented example of polymorphism is Labrador Retriever coloring; while the coat colour depends on many genes, it is clearly seen in the surroundings equally yellow, black, and brown. Richard Dawkins in 1978[3] and so again in his 1982 volume The Extended Phenotype suggested that i can regard bird nests and other built structures such equally caddis-fly larva cases and beaver dams every bit "extended phenotypes".
Wilhelm Johannsen proposed the genotype–phenotype distinction in 1911 to make clear the difference between an organism's heredity and what that heredity produces.[4] [5] The distinction resembles that proposed past August Weismann (1834–1914), who distinguished between germ plasm (heredity) and somatic cells (the body).
The genotype–phenotype distinction should not be confused with Francis Crick's primal dogma of molecular biology, a statement about the directionality of molecular sequential information flowing from DNA to protein, and not the reverse.
Difficulties in definition [edit]
Despite its seemingly straightforward definition, the concept of the phenotype has hidden subtleties. Information technology may seem that annihilation dependent on the genotype is a phenotype, including molecules such every bit RNA and proteins. Most molecules and structures coded by the genetic fabric are not visible in the appearance of an organism, yet they are observable (for example by Western blotting) and are thus role of the phenotype; human blood groups are an example. It may seem that this goes beyond the original intentions of the concept with its focus on the (living) organism in itself. Either way, the term phenotype includes inherent traits or characteristics that are observable or traits that tin can be made visible past some technical procedure. A notable extension to this idea is the presence of "organic molecules" or metabolites that are generated past organisms from chemical reactions of enzymes.

ABO blood groups determined through a Punnett square and displaying phenotypes and genotypes
The term "phenotype" has sometimes been incorrectly used every bit a shorthand for the phenotypic difference between a mutant and its wild type, which (if not significant) leads to the argument that a "mutation has no phenotype".[6]
Some other extension adds behavior to the phenotype, since behaviors are observable characteristics. Behavioral phenotypes include cognitive, personality, and behavioral patterns. Some behavioral phenotypes may narrate psychiatric disorders[vii] or syndromes.[viii] [nine]


B.betularia morpha carbonaria, the melanic form, illustrating discontinuous variation
Phenotypic variation [edit]
Phenotypic variation (due to underlying heritable genetic variation) is a fundamental prerequisite for evolution by natural selection. Information technology is the living organism as a whole that contributes (or non) to the adjacent generation, so natural option affects the genetic structure of a population indirectly via the contribution of phenotypes. Without phenotypic variation, there would be no development by natural choice.[10]
The interaction between genotype and phenotype has ofttimes been conceptualized by the following human relationship:
- genotype (G) + environment (East) → phenotype (P)
A more than nuanced version of the relationship is:
- genotype (G) + environment (Due east) + genotype & environment interactions (GE) → phenotype (P)
Genotypes ofttimes have much flexibility in the modification and expression of phenotypes; in many organisms these phenotypes are very unlike under varying environmental atmospheric condition (see ecophenotypic variation). The plant Hieracium umbellatum is plant growing in two dissimilar habitats in Sweden. One habitat is rocky, sea-side cliffs, where the plants are bushy with broad leaves and expanded inflorescences; the other is among sand dunes where the plants abound prostrate with narrow leaves and meaty inflorescences. These habitats alternate forth the coast of Sweden and the habitat that the seeds of Hieracium umbellatum land in, determine the phenotype that grows.[11]
An instance of random variation in Drosophila flies is the number of ommatidia, which may vary (randomly) between left and right eyes in a single private every bit much equally they do betwixt different genotypes overall, or between clones raised in dissimilar environments.[ citation needed ]
The concept of phenotype tin can be extended to variations below the level of the gene that affect an organism'south fitness. For example, silent mutations that do non change the corresponding amino acrid sequence of a gene may modify the frequency of guanine-cytosine base pairs (GC content). These base pairs take a higher thermal stability (melting point) than adenine-thymine, a property that might convey, among organisms living in high-temperature environments, a selective advantage on variants enriched in GC content.[ citation needed ]
The extended phenotype [edit]
Richard Dawkins described a phenotype that included all effects that a gene has on its surroundings, including other organisms, as an extended phenotype, arguing that "An animal'due south beliefs tends to maximize the survival of the genes 'for' that behavior, whether or not those genes happen to be in the body of the item animal performing it."[3] For case, an organism such as a beaver modifies its surroundings by building a beaver dam; this can exist considered an expression of its genes, just equally its incisor teeth are—which it uses to modify its environment. Similarly, when a bird feeds a brood parasite such as a cuckoo, it is unwittingly extending its phenotype; and when genes in an orchid bear on orchid bee behavior to increase pollination, or when genes in a peacock affect the copulatory decisions of peahens, again, the phenotype is being extended. Genes are, in Dawkins's view, selected past their phenotypic effects.[12]
Other biologists broadly hold that the extended phenotype concept is relevant, but consider that its office is largely explanatory, rather than assisting in the blueprint of experimental tests.[13]
Phenome and phenomics [edit]
Although a phenotype is the ensemble of observable characteristics displayed by an organism, the word phenome is sometimes used to refer to a collection of traits, while the simultaneous study of such a collection is referred to as phenomics.[14] [fifteen] Phenomics is an important field of study because information technology tin can be used to effigy out which genomic variants impact phenotypes which then tin be used to explain things like wellness, disease, and evolutionary fitness.[sixteen] Phenomics forms a big role of the Human Genome Projection[17]
Phenomics has applications in agriculture. For example, genomic variations such every bit drought and heat resistance can be identified through phenomics to create more durable GMOs.[xviii] [nineteen]
Phenomics may be a stepping stone towards personalized medicine, peculiarly drug therapy.[20] Once the phenomic database has acquired more than information, a person's phenomic information tin be used to select specific drugs tailored to an individual.[twenty]
Large-calibration phenotyping and genetic screens [edit]
Large-scale genetic screens can identify the genes or mutations that bear on the phenotype of an organism. Analyzing the phenotypes of mutant genes can likewise help in determining factor function.[21] For example, a large-scale phenotypic screen has been used to study lesser understood phenotypes such as beliefs. In this screen, the office of mutations in mice were studied in areas such equally learning and memory, circadian rhythmicity, vision, responses to stress and response to psychostimulants (see table for details).
| Phenotypic Domain | Assay | Notes | Software Packet |
| Circadian Rhythm | Bike running behavior | ClockLab | |
| Learning and Retentivity | Fright conditioning | Video-image-based scoring of freezing | FreezeFrame |
| Preliminary Assessment | Open field activity and elevated plus maze | Video-image-based scoring of exploration | LimeLight |
| Psychostimulant response | Hyperlocomotion beliefs | Video-image-based tracking of locomotion | BigBrother |
| Vision | Electroretinogram and Fundus photography | L. Pinto and colleagues |
This experiment involved the progeny of mice treated with ENU, or N-ethyl-N-nitrosourea, which is a potent mutagen that causes betoken mutations. The mice were phenotypically screened for alterations in the different behavioral domains in guild to detect the number of putative mutants (come across table for details). Putative mutants are then tested for heritability in club to help determine the inheritance design as well every bit map out the mutations. Once they have been mapped out, cloned, and identified, it can be determined whether a mutation represents a new gene or not.
| Phenotypic domain | ENU Progeny screened | Putative mutants | Putative mutant lines with progeny | Confirmed mutants |
| General assessment | 29860 | fourscore | 38 | 14 |
| Learning and retentivity | 23123 | 165 | 106 | 19 |
| Psychostimulant response | 20997 | 168 | 86 | 9 |
| Neuroendocrine response to stress | 13118 | 126 | 54 | ii |
| Vision | 15582 | 108 | 60 | 6 |
These experiments showed that mutations in the rhodopsin gene afflicted vision and tin can even cause retinal degeneration in mice.[22] The aforementioned amino acid change causes homo familial incomprehension, showing how phenotyping in animals tin can inform medical diagnostics and mayhap therapy.
Evolutionary origin of phenotype [edit]
The RNA globe is the hypothesized pre-cellular phase in the evolutionary history of life on earth, in which self-replicating RNA molecules proliferated prior to the evolution of Dna and proteins.[23] The folded three-dimensional physical structure of the first RNA molecule that possessed ribozyme action promoting replication while fugitive destruction would take been the showtime phenotype, and the nucleotide sequence of the first cocky-replicating RNA molecule would have been the original genotype.[23]
See as well [edit]
- Ecotype
- Endophenotype
- Genotype
- Genotype-phenotype stardom
- Molecular phenotyping
- Race and genetics
References [edit]
- ^ "phenotype adjective – Definition, pictures, pronunciation and usage notes". Oxford Advanced Learner's Dictionary at OxfordLearnersDictionaries.com . Retrieved 2020-04-29 .
the ready of observable characteristics of an private resulting from the interaction of its genotype with the environment.
- ^ "Genotype versus phenotype". Understanding Evolution . Retrieved 2020-04-29 .
An organism'south genotype is the set of genes that it carries. An organism'southward phenotype is all of its appreciable characteristics — which are influenced both by its genotype and by the environment.
- ^ a b Dawkins R (May 1978). "Replicator choice and the extended phenotype". Zeitschrift Fur Tierpsychologie. 47 (1): 61–76. doi:10.1111/j.1439-0310.1978.tb01823.x. PMID 696023.
- ^ Churchill FB (1974). "William Johannsen and the genotype concept". Periodical of the History of Biology. 7 (1): v–30. doi:x.1007/BF00179291. PMID 11610096. S2CID 38649212.
- ^ Johannsen W (August 2014). "The genotype conception of heredity. 1911". International Periodical of Epidemiology. 43 (4): 989–yard. doi:10.1086/279202. JSTOR 2455747. PMC4258772. PMID 24691957.
- ^ Crusio Nosotros (May 2002). "'My mouse has no phenotype'". Genes, Brain, and Behavior. 1 (2): 71. doi:ten.1034/j.1601-183X.2002.10201.10. PMID 12884976. S2CID 35382304.
- ^ Cassidy SB, Morris CA (2002-01-01). "Behavioral phenotypes in genetic syndromes: genetic clues to human being behavior". Advances in Pediatrics. 49: 59–86. PMID 12214780.
- ^ O'Brien G, Yule W, eds. (1995). Behavioural Phenotype. Clinics in Developmental Medicine No.138. London: Mac Keith Printing. ISBN978-1-898683-06-3.
- ^ O'Brien, Gregory, ed. (2002). Behavioural Phenotypes in Clinical Practice. London: Mac Keith Printing. ISBN978-1-898683-27-eight . Retrieved 27 September 2010.
- ^ Lewontin RC (November 1970). "The Units of Selection" (PDF). Annual Review of Environmental and Systematics. 1: one–18. doi:10.1146/annurev.es.01.110170.000245. JSTOR 2096764.
- ^ "Botany online: Evolution: The Modern Synthesis - Phenotypic and Genetic Variation; Ecotypes". Archived from the original on 2009-06-18. Retrieved 2009-12-29 .
- ^ Dawkins R (1982). The Extended Phenotype. Oxford University. p. iv. ISBN978-0-19-288051-2.
- ^ Hunter P (March 2009). "Extended phenotype redux. How far can the accomplish of genes extend in manipulating the environment of an organism?". EMBO Reports. 10 (iii): 212–215. doi:10.1038/embor.2009.xviii. PMC2658563. PMID 19255576.
- ^ Mahner Thou, Kary M (May 1997). "What exactly are genomes, genotypes and phenotypes? And what virtually phenomes?". Periodical of Theoretical Biology. 186 (ane): 55–63. Bibcode:1997JThBi.186...55M. doi:x.1006/jtbi.1996.0335. PMID 9176637.
- ^ Varki A, Wills C, Perlmutter D, Woodruff D, Cuff F, Moore J, et al. (October 1998). "Not bad Ape Phenome Project?". Scientific discipline. 282 (5387): 239–240. Bibcode:1998Sci...282..239V. doi:10.1126/scientific discipline.282.5387.239d. PMID 9841385. S2CID 5837659.
- ^ Houle D, Govindaraju DR, Omholt S (Dec 2010). "Phenomics: the next challenge". Nature Reviews. Genetics. 11 (12): 855–866. doi:ten.1038/nrg2897. PMID 21085204. S2CID 14752610.
- ^ Freimer N, Sabatti C (May 2003). "The homo phenome project". Nature Genetics. 34 (1): 15–21. doi:ten.1038/ng0503-xv. PMID 12721547. S2CID 31510391.
- ^ Rahman H, Ramanathan V, Jagadeeshselvam N, Ramasamy S, Rajendran S, Ramachandran M, et al. (2015-01-01). "Phenomics: technologies and applications in plant and agronomics.". In Barh D, Khan MS, Davies Due east (eds.). PlantOmics: The Omics of Plant Science. New Delhi: Springer. pp. 385–411. doi:10.1007/978-81-322-2172-2_13. ISBN9788132221715.
- ^ Furbank RT, Tester G (December 2011). "Phenomics--technologies to salve the phenotyping bottleneck". Trends in Plant Science. xvi (12): 635–644. doi:ten.1016/j.tplants.2011.09.005. PMID 22074787.
- ^ a b Monte AA, Brocker C, Nebert DW, Gonzalez FJ, Thompson DC, Vasiliou V (September 2014). "Improved drug therapy: triangulating phenomics with genomics and metabolomics". Human Genomics. 8 (1): 16. doi:x.1186/s40246-014-0016-nine. PMC4445687. PMID 25181945.
- ^ Amsterdam A, Burgess S, Golling G, Chen W, Dominicus Z, Townsend Chiliad, et al. (Oct 1999). "A large-scale insertional mutagenesis screen in zebrafish". Genes & Development. 13 (twenty): 2713–2724. doi:10.1101/gad.13.20.2713. PMC317115. PMID 10541557.
- ^ Vitaterna MH, Pinto LH, Takahashi JS (Apr 2006). "Large-scale mutagenesis and phenotypic screens for the nervous arrangement and beliefs in mice". Trends in Neurosciences. 29 (4): 233–240. doi:10.1016/j.tins.2006.02.006. PMC3761413. PMID 16519954.
- ^ a b Michod R (1983) Population biology of the first replicators: On the origin of the genotype, phenotype and organism. Am Zool 23:5–xiv
External links [edit]
| | Wikimedia Commons has media related to Phenotypes. |
- Mouse Phenome Database
- Human Phenotype Ontology
- Europhenome: Access to raw and annotated mouse phenotype information
- "Wilhelm Johannsen's Genotype-Phenotype Distinction" by E. Peirson at the Embryo Project Encyclopedia
Source: https://en.wikipedia.org/wiki/Phenotype
0 Response to "What Name Is Given to the Collection of Traits Exhibited by an Organism?"
Post a Comment